0$,
# the gamma distribution is the following,
#
# $$
# f(x;\alpha,\beta)=
# \frac{\beta ^{-\alpha } x^{\alpha
# -1}
# e^{-\frac{x}{\beta
# }}}{\Gamma (\alpha )}
# $$
#
# and $f(x;\alpha,\beta)=0$ when $x\le 0$ and $\Gamma$ is the gamma
# function. For
# example, suppose that vehicles passing under a gate follows a
# Poisson process,
# with an average of 5 vehicles passing per hour, what is the
# probability that at
# most an hour will have passed before 2 vehicles pass the
# gate? If $X$ is time in
# hours that transpires before the 2 vehicles pass, then
# we have $\beta=1/5$ and
# $\alpha=2$. The required probability $\mathbb{P}(X<1)
# \approx 0.96 $. The gamma
# distribution has $\mathbb{E}(X) = \alpha\beta $ and
# $\mathbb{V}(X)=\alpha\beta^2$
#
# The
# following code computes the result of the
# example above. Note that the
# parameters are described in slightly different
# terms as above, as described in
# the corresponding documentation for `gamma`.
# In[33]:
from scipy.stats import gamma
x = gamma(2,scale=1/5) # create random variable object
print(x.cdf(1))
# ## Beta Distribution
#
#
#
# The uniform distribution assigns a single constant value
# over the unit interval. The Beta distribution generalizes this to
# a function
# over the unit interval. The probability density function
# of the Beta
# distribution is the following,
#
# $$
# f(x ) = \frac{1}{\beta(a,b)} x^{a-1} (1-x)^{b-1}
# $$
#
# where
#
# $$
# \beta(a,b) = \int_0^1 x^{a-1} (1-x)^{b-1} dx
# $$
#
# Note that $a=b=1$ yields the uniform distribution. In the
# special case for
# integers where $0\le k\le n$, we have
#
# $$
# \int_0^1 \binom{n}{k}x^k (1-x)^{n-k} dx = \frac{1}{n+1}
# $$
#
# To get this result without calculus, we can use an experiment by
# Thomas Bayes.
# Start with $n$ white balls and one gray ball. Uniformly at
# random, toss them
# onto the unit interval. Let $X$ be the number of white balls
# to the left of the
# gray ball. Thus, $X\in \lbrace 0,1,\ldots,n \rbrace$. To
# compute
# $\mathbb{P}(X=k)$, we condition on the probability of the position $B$
# of the
# gray ball, which is uniformly distributed over the unit interval
# ($f(p)=1$).
# Thus, we have
#
# $$
# \mathbb{P}(X=k) = \int_0^1 \mathbb{P}(X=k\vert B=p) f(p) dp = \int_0^1
# \binom{n}{k}p^k (1-p)^{n-k} dp
# $$
#
# Now, consider a slight variation on the experiment where we start
# with $n+1$
# white balls and again toss them onto the unit interval and then
# later choose one
# ball at random to color gray. Using the same $X$ as before, by
# symmetry, because
# any one of the $n+1$ balls is equally likely to be chosen, we
# have
#
# $$
# \mathbb{P}(X=k)=\frac{1}{n+1}
# $$
#
# for $k\in \lbrace 0,1,\ldots,n \rbrace$. Both situations describe the
# same
# problem because it does not matter whether we paint the ball before or
# after we
# throw it. Setting the last two equations equal gives the desired
# result without
# using calculus.
#
# $$
# \int_0^1 \binom{n}{k}p^k (1-p)^{n-k} dp = \frac{1}{n+1}
# $$
#
# The following code shows where to get the Beta distribution from the `scipy`
# module.
# In[34]:
from scipy.stats import beta
x = beta(1,1) # create random variable object
print(x.cdf(1))
# Given this experiment, it is not too surprising that there is an intimate
# relationship between the Beta distribution and binomial random variables.
# Suppose we want to estimate the probability of heads for coin-tosses using
# Bayesian inference. Using this approach, all unknown quantities are treated as
# random variables. In this case, the probability of heads ($p$) is the unknown
# quantity that requires a *prior* distribution. Let us choose the Beta
# distribution as the prior distribution, $\texttt{Beta}(a,b)$. Then,
# conditioning
# on $p$, we have
#
# $$
# X\vert p \sim \texttt{binom}(n,p)
# $$
#
# which says that $X$ is conditionally distributed as a binomial. To
# get the
# posterior probability, $f(p\vert X=k)$, we have the following
# Bayes rule,
#
# $$
# f(p\vert X=k) = \frac{\mathbb{P}(X=k\vert p)f(p)}{\mathbb{P}(X=k)}
# $$
#
# with the corresponding denominator,
#
# $$
# \mathbb{P}(X=k) = \int_0^1 \binom{n}{k}p^k (1-p)^{n-k}f(p) dp
# $$
#
# Note that unlike with our experiment before, $f(p)$ is not constant.
# Without
# substituting in all of the distributions, we observe that the
# posterior is a
# function of $p$ which means that everything else that is not a
# function of $p$
# is a constant. This gives,
#
# $$
# f(p\vert X=k) \propto p^{a+k-1} (1-p)^{b+n-k-1}
# $$
#
# which is another Beta distribution with parameters $a+k,b+n-k$. This
# special
# relationship in which the beta prior probability distribution on $p$ on
# data
# that are conditionally binomial distributed yields the posterior that is
# also
# binomial distributed is known as *conjugacy*. We say that the Beta
# distribution
# is the conjugate prior of the binomial distribution.
#
# ## Dirichlet-multinomial
# Distribution
#
#
#
# The Dirichlet-multinomial distribution is a discrete
# multivariate distribution
# also known as the multivariate Polya distribution.
# The Dirichlet-multinomial
# distribution arises in situations where the usual
# multinomial distribution is
# inadequate. For example, if a multinomial
# distribution is used to model the
# number of balls that land in a set of bins and
# the multinomial parameter vector
# (i.e., probabilities of balls landing in
# particular bins) varies from trial to
# trial, then the Dirichlet distribution can
# be used to include variation in
# those probabilities because the Dirichlet
# distribution is defined over a
# simplex that describes the multinomial parameter
# vector.
#
# Specifically, suppose we have $K$ rival events, each with probability
# $\mu_k$.
# Then, the probability of the vector $\boldsymbol{\mu}$ given that
# each
# event has been observed $\alpha_k$ times is the following,
#
# $$
# \mathbb{P}(\boldsymbol{\mu}\vert \boldsymbol{\alpha}) \propto \prod_{k=1}^K
# \mu_k^{\alpha_k-1}
# $$
#
# where $0\le\mu_k\le 1$ and $\sum\mu_k=1$. Note that this last sum is
# a
# constraint that makes the distribution $K-1$ dimensional. The normalizing
# constant for this distribution is the multinomial Beta function,
#
# $$
# \texttt{Beta}(\boldsymbol{\alpha})=\frac{\prod_{k=1}^K\Gamma(\alpha_k)}{\Gamma(\sum_{k=1}^K\alpha_k)}
# $$
#
# The elements of the $\boldsymbol{\alpha}$ vector are also called
# *concentration* parameters. As before, the Dirichlet
# distribution can be found
# in the `scipy.stats` module,
# In[35]:
from scipy.stats import dirichlet
d = dirichlet([ 1,1,1 ])
d.rvs(3) # get samples from distribution
# Note that each of the rows sums to one. This is because of the
# $\sum\mu_k=1$
# constraint. We can generate more samples and plot this
# using `Axes3D` in
# Matplotlib in [Figure](#fig:Dirichlet_001).
# In[36]:
get_ipython().run_line_magic('matplotlib', 'inline')
from mpl_toolkits.mplot3d import Axes3D
from matplotlib.pyplot import subplots
x = d.rvs(1000)
fig, ax = subplots(subplot_kw=dict(projection='3d'))
_=ax.scatter(x[:,0],x[:,1],x[:,2],marker='o',alpha=.2)
ax.view_init(30, 30) # elevation, azimuth
# ax.set_aspect(1)
_=ax.set_xlabel(r'$\mu_1$')
_=ax.set_ylabel(r'$\mu_2$')
_=ax.set_zlabel(r'$\mu_3$')
fig.savefig('fig-probability/Dirichlet_001.png')
#
#
#
#
# One thousand samples from a Dirichlet
# distribution with $\boldsymbol{\alpha} = [1,1,1]$
#  #
#
#
#
# Notice that the
# generated samples lie on the triangular simplex shown. The
# corners of the
# triangle correspond to each of the components in the
# $\boldsymbol{\mu}$. Using,
# a non-uniform $\boldsymbol{\alpha}=[2,3,4]$ vector,
# we can visualize the
# probability density function using the `pdf` method on the
# `dirichlet` object as
# shown in [Figure](#fig:Dirichlet_002). By choosing the
# $\boldsymbol{\alpha}\in
# \mathbb{R}^3$, the peak of the density function can be
# moved within the
# corresponding triangular simplex.
# In[37]:
import numpy as np
from matplotlib.pylab import cm
X,Y = np.meshgrid(np.linspace(.01,1,50),np.linspace(.01,1,50))
d = dirichlet([2,3,4])
idx=(X+Y<1)
f=d.pdf(np.vstack([X[idx],Y[idx],1-X[idx]-Y[idx]]))
Z = idx*0+ np.nan
Z[idx] = f
fig,ax=subplots()
_=ax.contourf(X,Y,Z,cmap=cm.viridis)
ax.set_aspect(1)
_=ax.set_xlabel('x',fontsize=18)
_=ax.set_ylabel('y',fontsize=18)
fig.savefig('fig-probability/Dirichlet_002.png')
#
#
#
#
#
#
#
#
#
# Notice that the
# generated samples lie on the triangular simplex shown. The
# corners of the
# triangle correspond to each of the components in the
# $\boldsymbol{\mu}$. Using,
# a non-uniform $\boldsymbol{\alpha}=[2,3,4]$ vector,
# we can visualize the
# probability density function using the `pdf` method on the
# `dirichlet` object as
# shown in [Figure](#fig:Dirichlet_002). By choosing the
# $\boldsymbol{\alpha}\in
# \mathbb{R}^3$, the peak of the density function can be
# moved within the
# corresponding triangular simplex.
# In[37]:
import numpy as np
from matplotlib.pylab import cm
X,Y = np.meshgrid(np.linspace(.01,1,50),np.linspace(.01,1,50))
d = dirichlet([2,3,4])
idx=(X+Y<1)
f=d.pdf(np.vstack([X[idx],Y[idx],1-X[idx]-Y[idx]]))
Z = idx*0+ np.nan
Z[idx] = f
fig,ax=subplots()
_=ax.contourf(X,Y,Z,cmap=cm.viridis)
ax.set_aspect(1)
_=ax.set_xlabel('x',fontsize=18)
_=ax.set_ylabel('y',fontsize=18)
fig.savefig('fig-probability/Dirichlet_002.png')
#
#
#
#
# Probability density function
# for the Dirichlet distribution with $\boldsymbol{\alpha}=[2,3,4]$
# 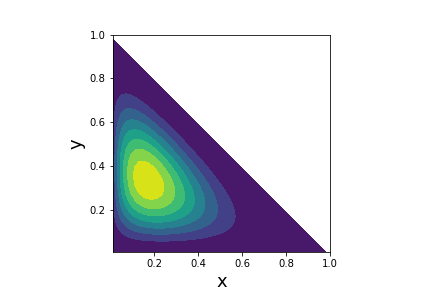 #
#
#
#
# We
# have seen that the Beta distribution generalizes the uniform distribution
# over
# the unit interval. Likewise, the Dirichlet distribution generalizes the
# Beta
# distribution over a vector with components in the unit interval. Recall
# that
# binomial distribution and the Beta distribution form a conjugate pair
# for
# Bayesian inference because with $p\sim \texttt{Beta} $,
#
# $$
# X\vert p \sim \texttt{Binomial}(n,p)
# $$
#
# That is, the data conditioned on $p$, is binomial distributed.
# Analogously, the
# multinomial distribution and the Dirichlet distribution also
# form such a
# conjugate pair with multinomial parameter $p\sim
# \texttt{Dirichlet} $,
#
# $$
# X\vert p \sim \texttt{multinomial}(n,p)
# $$
#
# For this reason, the Dirichlet-multinomial distribution is popular in
# machine
# learning text processing because non-zero probabilities can be assigned
# to words
# not specifically contained in specific documents, which helps
# generalization
# performance.
#
#
# ## Negative Binomial Distribution
#
#
# The negative binomial
# distribution is used to characterize the number
# of trials until a specified
# number of failures ($r$) occurs. For
# example, suppose `1` indicates failure
# and `0` indicates success. Then
# the negative binomial distribution characterizes
# the probability of a
# `k=6` long sequence that has two (`r=2`) failures, with the
# sequence
# terminating in a failure (e.g., `001001`) with
# $\mathbb{P}(1)=1/3$.
# The length of the sequence is `6`, so for the
# negative binomial distribution,
# $\mathbb{P}(6-2)=\frac{80}{729}$.
#
# The probability mass function is the
# following:
#
# $$
# \mathbb{P}(k) = \binom{n+k-1}{n-1} p^n (1-p)^k
# $$
#
# where $p$ is the probability of failure. The mean and
# variance of this
# distribution is the following:
#
# $$
# \mathbb{E}(k) =\frac{n (1-p)}{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n (1-p)}{p^2}
# $$
#
# The following simulation shows an example
# sequence generated for the negative
# binomial distribution.
# In[38]:
import random
n=2 # num of failures
p=1/3 # prob of failure
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices([0,1],[1-p,p])
seq.append(v)
nc += (v == 1)
seq,len(seq)
# Keep in mind that the negative binomial distribution characterizes
# the family
# of such sequences with the specified number of failures.
#
# ## Negative
# Multinomial Distribution
#
#
# The discrete negative multinomial distribution is an
# extension of the negative
# binomial distribution to account for more than two
# possible outcomes. That is,
# there are other alteratives whose respective
# probabilities sum to one less the
# failure probability, $p_{f} = 1-\sum_{k=1}^n
# p_i$. For example, a random
# sample from this distribution with parameters $n=2$
# (number of observed
# failures) and with $p_a= \frac{1}{3}, p_b=\frac{1}{2}$ means
# that the failure
# probability, $p_f=\frac{1}{6}$. Thus, a sample from this
# distribution like
# $[ 2,9]$ means that `2` of the $a$ objects were observed in
# the
# sequence, `9` of the $b$ objects were observed, and there were two failure
# symbols (say, `F`) with one of them at the end of the sequence.
#
# The probability
# mass function is the following:
#
# $$
# \mathbb{P}(\mathbf{k})= (n)_{\sum_{i=0}^m k_i} p_f^{n} \prod_{i=1}^m
# \frac{p_i^{k_i}}{k_i!}
# $$
#
# where $p_f$ is the probability of failure and the other $p_i$ terms
# are the
# probabilities of the other alternatives in the sequence. The
# $(a)_n$ notation
# is the rising factorial function (e.g., $a_3 = a (a+1)(a+2)$).
# The mean and
# variance of this distribution is the
# following:
#
# $$
# \mathbb{E}(\mathbf{k}) =\frac{n}{p_f} \mathbf{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n}{p_f^2} \mathbf{p} \mathbf{p}^T +
# \frac{n}{p_f}\diag(\mathbf{p})
# $$
#
# The following simulation shows the sequences generated for the
# negative
# multinomial distribution.
# In[39]:
import random
from collections import Counter
n=2 # num of failure items
p=[1/3,1/2] # prob of other non-failure items
items = ['a','b','F'] # F marks failure item
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices(items,p+[1-sum(p)])
seq.append(v)
nc += (v == 'F')
c=Counter(seq)
print(c)
# The values of the `Counter` dictionary above are the $\mathbf{k}$
# vectors in
# the probability mass function for the negative multinomial distribution.
# Importantly, these are not the probabilities of a particular sequence, but
# of a
# family of sequences with the same corresponding `Counter` values.
# The
# probability mass function implemented in Python is the following,
# In[40]:
from scipy.special import factorial
import numpy as np
def negative_multinom_pdf(p,n):
assert len(n) == len(p)
term = [i**j for i,j in zip(p,n)]
num=np.prod(term)*(1-sum(p))*factorial(sum(n))
den = np.prod([factorial(i) for i in n])
return num/den
# Evaluating this with the prior `Counter` result,
# In[41]:
negative_multinom_pdf([1/3,1/2],[c['a'],c['b']])
#
#
#
#
# We
# have seen that the Beta distribution generalizes the uniform distribution
# over
# the unit interval. Likewise, the Dirichlet distribution generalizes the
# Beta
# distribution over a vector with components in the unit interval. Recall
# that
# binomial distribution and the Beta distribution form a conjugate pair
# for
# Bayesian inference because with $p\sim \texttt{Beta} $,
#
# $$
# X\vert p \sim \texttt{Binomial}(n,p)
# $$
#
# That is, the data conditioned on $p$, is binomial distributed.
# Analogously, the
# multinomial distribution and the Dirichlet distribution also
# form such a
# conjugate pair with multinomial parameter $p\sim
# \texttt{Dirichlet} $,
#
# $$
# X\vert p \sim \texttt{multinomial}(n,p)
# $$
#
# For this reason, the Dirichlet-multinomial distribution is popular in
# machine
# learning text processing because non-zero probabilities can be assigned
# to words
# not specifically contained in specific documents, which helps
# generalization
# performance.
#
#
# ## Negative Binomial Distribution
#
#
# The negative binomial
# distribution is used to characterize the number
# of trials until a specified
# number of failures ($r$) occurs. For
# example, suppose `1` indicates failure
# and `0` indicates success. Then
# the negative binomial distribution characterizes
# the probability of a
# `k=6` long sequence that has two (`r=2`) failures, with the
# sequence
# terminating in a failure (e.g., `001001`) with
# $\mathbb{P}(1)=1/3$.
# The length of the sequence is `6`, so for the
# negative binomial distribution,
# $\mathbb{P}(6-2)=\frac{80}{729}$.
#
# The probability mass function is the
# following:
#
# $$
# \mathbb{P}(k) = \binom{n+k-1}{n-1} p^n (1-p)^k
# $$
#
# where $p$ is the probability of failure. The mean and
# variance of this
# distribution is the following:
#
# $$
# \mathbb{E}(k) =\frac{n (1-p)}{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n (1-p)}{p^2}
# $$
#
# The following simulation shows an example
# sequence generated for the negative
# binomial distribution.
# In[38]:
import random
n=2 # num of failures
p=1/3 # prob of failure
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices([0,1],[1-p,p])
seq.append(v)
nc += (v == 1)
seq,len(seq)
# Keep in mind that the negative binomial distribution characterizes
# the family
# of such sequences with the specified number of failures.
#
# ## Negative
# Multinomial Distribution
#
#
# The discrete negative multinomial distribution is an
# extension of the negative
# binomial distribution to account for more than two
# possible outcomes. That is,
# there are other alteratives whose respective
# probabilities sum to one less the
# failure probability, $p_{f} = 1-\sum_{k=1}^n
# p_i$. For example, a random
# sample from this distribution with parameters $n=2$
# (number of observed
# failures) and with $p_a= \frac{1}{3}, p_b=\frac{1}{2}$ means
# that the failure
# probability, $p_f=\frac{1}{6}$. Thus, a sample from this
# distribution like
# $[ 2,9]$ means that `2` of the $a$ objects were observed in
# the
# sequence, `9` of the $b$ objects were observed, and there were two failure
# symbols (say, `F`) with one of them at the end of the sequence.
#
# The probability
# mass function is the following:
#
# $$
# \mathbb{P}(\mathbf{k})= (n)_{\sum_{i=0}^m k_i} p_f^{n} \prod_{i=1}^m
# \frac{p_i^{k_i}}{k_i!}
# $$
#
# where $p_f$ is the probability of failure and the other $p_i$ terms
# are the
# probabilities of the other alternatives in the sequence. The
# $(a)_n$ notation
# is the rising factorial function (e.g., $a_3 = a (a+1)(a+2)$).
# The mean and
# variance of this distribution is the
# following:
#
# $$
# \mathbb{E}(\mathbf{k}) =\frac{n}{p_f} \mathbf{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n}{p_f^2} \mathbf{p} \mathbf{p}^T +
# \frac{n}{p_f}\diag(\mathbf{p})
# $$
#
# The following simulation shows the sequences generated for the
# negative
# multinomial distribution.
# In[39]:
import random
from collections import Counter
n=2 # num of failure items
p=[1/3,1/2] # prob of other non-failure items
items = ['a','b','F'] # F marks failure item
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices(items,p+[1-sum(p)])
seq.append(v)
nc += (v == 'F')
c=Counter(seq)
print(c)
# The values of the `Counter` dictionary above are the $\mathbf{k}$
# vectors in
# the probability mass function for the negative multinomial distribution.
# Importantly, these are not the probabilities of a particular sequence, but
# of a
# family of sequences with the same corresponding `Counter` values.
# The
# probability mass function implemented in Python is the following,
# In[40]:
from scipy.special import factorial
import numpy as np
def negative_multinom_pdf(p,n):
assert len(n) == len(p)
term = [i**j for i,j in zip(p,n)]
num=np.prod(term)*(1-sum(p))*factorial(sum(n))
den = np.prod([factorial(i) for i in n])
return num/den
# Evaluating this with the prior `Counter` result,
# In[41]:
negative_multinom_pdf([1/3,1/2],[c['a'],c['b']])
 #
#
#
#
# Notice that the
# generated samples lie on the triangular simplex shown. The
# corners of the
# triangle correspond to each of the components in the
# $\boldsymbol{\mu}$. Using,
# a non-uniform $\boldsymbol{\alpha}=[2,3,4]$ vector,
# we can visualize the
# probability density function using the `pdf` method on the
# `dirichlet` object as
# shown in [Figure](#fig:Dirichlet_002). By choosing the
# $\boldsymbol{\alpha}\in
# \mathbb{R}^3$, the peak of the density function can be
# moved within the
# corresponding triangular simplex.
# In[37]:
import numpy as np
from matplotlib.pylab import cm
X,Y = np.meshgrid(np.linspace(.01,1,50),np.linspace(.01,1,50))
d = dirichlet([2,3,4])
idx=(X+Y<1)
f=d.pdf(np.vstack([X[idx],Y[idx],1-X[idx]-Y[idx]]))
Z = idx*0+ np.nan
Z[idx] = f
fig,ax=subplots()
_=ax.contourf(X,Y,Z,cmap=cm.viridis)
ax.set_aspect(1)
_=ax.set_xlabel('x',fontsize=18)
_=ax.set_ylabel('y',fontsize=18)
fig.savefig('fig-probability/Dirichlet_002.png')
#
#
#
#
#
#
#
#
#
# Notice that the
# generated samples lie on the triangular simplex shown. The
# corners of the
# triangle correspond to each of the components in the
# $\boldsymbol{\mu}$. Using,
# a non-uniform $\boldsymbol{\alpha}=[2,3,4]$ vector,
# we can visualize the
# probability density function using the `pdf` method on the
# `dirichlet` object as
# shown in [Figure](#fig:Dirichlet_002). By choosing the
# $\boldsymbol{\alpha}\in
# \mathbb{R}^3$, the peak of the density function can be
# moved within the
# corresponding triangular simplex.
# In[37]:
import numpy as np
from matplotlib.pylab import cm
X,Y = np.meshgrid(np.linspace(.01,1,50),np.linspace(.01,1,50))
d = dirichlet([2,3,4])
idx=(X+Y<1)
f=d.pdf(np.vstack([X[idx],Y[idx],1-X[idx]-Y[idx]]))
Z = idx*0+ np.nan
Z[idx] = f
fig,ax=subplots()
_=ax.contourf(X,Y,Z,cmap=cm.viridis)
ax.set_aspect(1)
_=ax.set_xlabel('x',fontsize=18)
_=ax.set_ylabel('y',fontsize=18)
fig.savefig('fig-probability/Dirichlet_002.png')
#
#
#
#
#  #
#
#
#
# We
# have seen that the Beta distribution generalizes the uniform distribution
# over
# the unit interval. Likewise, the Dirichlet distribution generalizes the
# Beta
# distribution over a vector with components in the unit interval. Recall
# that
# binomial distribution and the Beta distribution form a conjugate pair
# for
# Bayesian inference because with $p\sim \texttt{Beta} $,
#
# $$
# X\vert p \sim \texttt{Binomial}(n,p)
# $$
#
# That is, the data conditioned on $p$, is binomial distributed.
# Analogously, the
# multinomial distribution and the Dirichlet distribution also
# form such a
# conjugate pair with multinomial parameter $p\sim
# \texttt{Dirichlet} $,
#
# $$
# X\vert p \sim \texttt{multinomial}(n,p)
# $$
#
# For this reason, the Dirichlet-multinomial distribution is popular in
# machine
# learning text processing because non-zero probabilities can be assigned
# to words
# not specifically contained in specific documents, which helps
# generalization
# performance.
#
#
# ## Negative Binomial Distribution
#
#
# The negative binomial
# distribution is used to characterize the number
# of trials until a specified
# number of failures ($r$) occurs. For
# example, suppose `1` indicates failure
# and `0` indicates success. Then
# the negative binomial distribution characterizes
# the probability of a
# `k=6` long sequence that has two (`r=2`) failures, with the
# sequence
# terminating in a failure (e.g., `001001`) with
# $\mathbb{P}(1)=1/3$.
# The length of the sequence is `6`, so for the
# negative binomial distribution,
# $\mathbb{P}(6-2)=\frac{80}{729}$.
#
# The probability mass function is the
# following:
#
# $$
# \mathbb{P}(k) = \binom{n+k-1}{n-1} p^n (1-p)^k
# $$
#
# where $p$ is the probability of failure. The mean and
# variance of this
# distribution is the following:
#
# $$
# \mathbb{E}(k) =\frac{n (1-p)}{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n (1-p)}{p^2}
# $$
#
# The following simulation shows an example
# sequence generated for the negative
# binomial distribution.
# In[38]:
import random
n=2 # num of failures
p=1/3 # prob of failure
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices([0,1],[1-p,p])
seq.append(v)
nc += (v == 1)
seq,len(seq)
# Keep in mind that the negative binomial distribution characterizes
# the family
# of such sequences with the specified number of failures.
#
# ## Negative
# Multinomial Distribution
#
#
# The discrete negative multinomial distribution is an
# extension of the negative
# binomial distribution to account for more than two
# possible outcomes. That is,
# there are other alteratives whose respective
# probabilities sum to one less the
# failure probability, $p_{f} = 1-\sum_{k=1}^n
# p_i$. For example, a random
# sample from this distribution with parameters $n=2$
# (number of observed
# failures) and with $p_a= \frac{1}{3}, p_b=\frac{1}{2}$ means
# that the failure
# probability, $p_f=\frac{1}{6}$. Thus, a sample from this
# distribution like
# $[ 2,9]$ means that `2` of the $a$ objects were observed in
# the
# sequence, `9` of the $b$ objects were observed, and there were two failure
# symbols (say, `F`) with one of them at the end of the sequence.
#
# The probability
# mass function is the following:
#
# $$
# \mathbb{P}(\mathbf{k})= (n)_{\sum_{i=0}^m k_i} p_f^{n} \prod_{i=1}^m
# \frac{p_i^{k_i}}{k_i!}
# $$
#
# where $p_f$ is the probability of failure and the other $p_i$ terms
# are the
# probabilities of the other alternatives in the sequence. The
# $(a)_n$ notation
# is the rising factorial function (e.g., $a_3 = a (a+1)(a+2)$).
# The mean and
# variance of this distribution is the
# following:
#
# $$
# \mathbb{E}(\mathbf{k}) =\frac{n}{p_f} \mathbf{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n}{p_f^2} \mathbf{p} \mathbf{p}^T +
# \frac{n}{p_f}\diag(\mathbf{p})
# $$
#
# The following simulation shows the sequences generated for the
# negative
# multinomial distribution.
# In[39]:
import random
from collections import Counter
n=2 # num of failure items
p=[1/3,1/2] # prob of other non-failure items
items = ['a','b','F'] # F marks failure item
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices(items,p+[1-sum(p)])
seq.append(v)
nc += (v == 'F')
c=Counter(seq)
print(c)
# The values of the `Counter` dictionary above are the $\mathbf{k}$
# vectors in
# the probability mass function for the negative multinomial distribution.
# Importantly, these are not the probabilities of a particular sequence, but
# of a
# family of sequences with the same corresponding `Counter` values.
# The
# probability mass function implemented in Python is the following,
# In[40]:
from scipy.special import factorial
import numpy as np
def negative_multinom_pdf(p,n):
assert len(n) == len(p)
term = [i**j for i,j in zip(p,n)]
num=np.prod(term)*(1-sum(p))*factorial(sum(n))
den = np.prod([factorial(i) for i in n])
return num/den
# Evaluating this with the prior `Counter` result,
# In[41]:
negative_multinom_pdf([1/3,1/2],[c['a'],c['b']])
#
#
#
#
# We
# have seen that the Beta distribution generalizes the uniform distribution
# over
# the unit interval. Likewise, the Dirichlet distribution generalizes the
# Beta
# distribution over a vector with components in the unit interval. Recall
# that
# binomial distribution and the Beta distribution form a conjugate pair
# for
# Bayesian inference because with $p\sim \texttt{Beta} $,
#
# $$
# X\vert p \sim \texttt{Binomial}(n,p)
# $$
#
# That is, the data conditioned on $p$, is binomial distributed.
# Analogously, the
# multinomial distribution and the Dirichlet distribution also
# form such a
# conjugate pair with multinomial parameter $p\sim
# \texttt{Dirichlet} $,
#
# $$
# X\vert p \sim \texttt{multinomial}(n,p)
# $$
#
# For this reason, the Dirichlet-multinomial distribution is popular in
# machine
# learning text processing because non-zero probabilities can be assigned
# to words
# not specifically contained in specific documents, which helps
# generalization
# performance.
#
#
# ## Negative Binomial Distribution
#
#
# The negative binomial
# distribution is used to characterize the number
# of trials until a specified
# number of failures ($r$) occurs. For
# example, suppose `1` indicates failure
# and `0` indicates success. Then
# the negative binomial distribution characterizes
# the probability of a
# `k=6` long sequence that has two (`r=2`) failures, with the
# sequence
# terminating in a failure (e.g., `001001`) with
# $\mathbb{P}(1)=1/3$.
# The length of the sequence is `6`, so for the
# negative binomial distribution,
# $\mathbb{P}(6-2)=\frac{80}{729}$.
#
# The probability mass function is the
# following:
#
# $$
# \mathbb{P}(k) = \binom{n+k-1}{n-1} p^n (1-p)^k
# $$
#
# where $p$ is the probability of failure. The mean and
# variance of this
# distribution is the following:
#
# $$
# \mathbb{E}(k) =\frac{n (1-p)}{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n (1-p)}{p^2}
# $$
#
# The following simulation shows an example
# sequence generated for the negative
# binomial distribution.
# In[38]:
import random
n=2 # num of failures
p=1/3 # prob of failure
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices([0,1],[1-p,p])
seq.append(v)
nc += (v == 1)
seq,len(seq)
# Keep in mind that the negative binomial distribution characterizes
# the family
# of such sequences with the specified number of failures.
#
# ## Negative
# Multinomial Distribution
#
#
# The discrete negative multinomial distribution is an
# extension of the negative
# binomial distribution to account for more than two
# possible outcomes. That is,
# there are other alteratives whose respective
# probabilities sum to one less the
# failure probability, $p_{f} = 1-\sum_{k=1}^n
# p_i$. For example, a random
# sample from this distribution with parameters $n=2$
# (number of observed
# failures) and with $p_a= \frac{1}{3}, p_b=\frac{1}{2}$ means
# that the failure
# probability, $p_f=\frac{1}{6}$. Thus, a sample from this
# distribution like
# $[ 2,9]$ means that `2` of the $a$ objects were observed in
# the
# sequence, `9` of the $b$ objects were observed, and there were two failure
# symbols (say, `F`) with one of them at the end of the sequence.
#
# The probability
# mass function is the following:
#
# $$
# \mathbb{P}(\mathbf{k})= (n)_{\sum_{i=0}^m k_i} p_f^{n} \prod_{i=1}^m
# \frac{p_i^{k_i}}{k_i!}
# $$
#
# where $p_f$ is the probability of failure and the other $p_i$ terms
# are the
# probabilities of the other alternatives in the sequence. The
# $(a)_n$ notation
# is the rising factorial function (e.g., $a_3 = a (a+1)(a+2)$).
# The mean and
# variance of this distribution is the
# following:
#
# $$
# \mathbb{E}(\mathbf{k}) =\frac{n}{p_f} \mathbf{p}
# $$
#
# $$
# \mathbb{V}(k) = \frac{n}{p_f^2} \mathbf{p} \mathbf{p}^T +
# \frac{n}{p_f}\diag(\mathbf{p})
# $$
#
# The following simulation shows the sequences generated for the
# negative
# multinomial distribution.
# In[39]:
import random
from collections import Counter
n=2 # num of failure items
p=[1/3,1/2] # prob of other non-failure items
items = ['a','b','F'] # F marks failure item
nc = 0 # counter
seq= []
while nc< n:
v,=random.choices(items,p+[1-sum(p)])
seq.append(v)
nc += (v == 'F')
c=Counter(seq)
print(c)
# The values of the `Counter` dictionary above are the $\mathbf{k}$
# vectors in
# the probability mass function for the negative multinomial distribution.
# Importantly, these are not the probabilities of a particular sequence, but
# of a
# family of sequences with the same corresponding `Counter` values.
# The
# probability mass function implemented in Python is the following,
# In[40]:
from scipy.special import factorial
import numpy as np
def negative_multinom_pdf(p,n):
assert len(n) == len(p)
term = [i**j for i,j in zip(p,n)]
num=np.prod(term)*(1-sum(p))*factorial(sum(n))
den = np.prod([factorial(i) for i in n])
return num/den
# Evaluating this with the prior `Counter` result,
# In[41]:
negative_multinom_pdf([1/3,1/2],[c['a'],c['b']])